Stefania’s work on the chromatin remodeler CHD7 is published
November 2020
Stefania found that CHD7, which is frequently mutated in CHARGE syndrome, is a poly(ADP-ribose)-dependent chromatin remodeler that works at sites of DNA damage and aids repair. Similar findings were made in the lab of Haico van Attikum and so our labs joined forces to dissect, helped by fantastic collaborators, the role of CHD7 for genome integrity maintenance. Great concerted effort, published in Nature Communications.
Read more about this work
Abstract
Chromatin structure is dynamically reorganized at multiple levels in response to DNA double-strand breaks (DSBs). Yet, how the different steps of chromatin reorganization are coordinated in space and time to differentially regulate DNA repair pathways is insufficiently understood. Here, we identify the Chromodomain Helicase DNA Binding Protein 7 (CHD7), which is frequently mutated in CHARGE syndrome, as an integral component of the non-homologous end-joining (NHEJ) DSB repair pathway. Upon recruitment via PARP1-triggered chromatin remodeling, CHD7 stimulates further chromatin relaxation around DNA break sites and brings in HDAC1/2 for localized chromatin de-acetylation. This counteracts the CHD7-induced chromatin expansion, thereby ensuring temporally and spatially controlled ‘chromatin breathing’ upon DNA damage, which we demonstrate fosters efficient and accurate DSB repair by controlling Ku and LIG4/XRCC4 activities. Loss of CHD7-HDAC1/2-dependent cNHEJ reinforces 53BP1 assembly at the damaged chromatin and shifts DSB repair to mutagenic NHEJ, revealing a backup function of 53BP1 when cNHEJ fails.
Further reading:
Stefania’s work on the chromatin remodeler CHD7 is published
November 2020
Stefania found that CHD7, which is frequently mutated in CHARGE syndrome, is a poly(ADP-ribose)-dependent chromatin remodeler that works at sites of DNA damage and aids repair. Similar findings were made in the lab of Haico van Attikum and so our labs joined forces to dissect, helped by fantastic collaborators, the role of CHD7 for genome integrity maintenance. Great concerted effort, published in Nature Communications.
Read more about this work
Abstract
Chromatin structure is dynamically reorganized at multiple levels in response to DNA double-strand breaks (DSBs). Yet, how the different steps of chromatin reorganization are coordinated in space and time to differentially regulate DNA repair pathways is insufficiently understood. Here, we identify the Chromodomain Helicase DNA Binding Protein 7 (CHD7), which is frequently mutated in CHARGE syndrome, as an integral component of the non-homologous end-joining (NHEJ) DSB repair pathway. Upon recruitment via PARP1-triggered chromatin remodeling, CHD7 stimulates further chromatin relaxation around DNA break sites and brings in HDAC1/2 for localized chromatin de-acetylation. This counteracts the CHD7-induced chromatin expansion, thereby ensuring temporally and spatially controlled ‘chromatin breathing’ upon DNA damage, which we demonstrate fosters efficient and accurate DSB repair by controlling Ku and LIG4/XRCC4 activities. Loss of CHD7-HDAC1/2-dependent cNHEJ reinforces 53BP1 assembly at the damaged chromatin and shifts DSB repair to mutagenic NHEJ, revealing a backup function of 53BP1 when cNHEJ fails.
Further reading:
Merula, Kyra and Daniel join the lab
October 2020
Merula Stout starts as a new PhD student in the Molecular Life Sciences Program of the Life Science Zurich Graduate School, Kyra Kirschenbühler as new student assistant, and Daniel Adler starts as new technical assistant to support the lab. Welcome on board! More info here.
Marco’s paper on TRIP12-regulated PARP inhibitor efficiency is out
August 2020
Marco could show that the ubiquitin E3 ligase TRIP12, through its PAR-binding WWE domain, targets PARP1 for proteasomal degradation, affecting PARP functions and PARP inhibitor efficiency. Great work with important implications worth following up. Congratulations to Marco and big thanks to our collaborators Qingyao Huang and Michael Baudis for their help. Published in Cell Reports.
Read more about Marco’s publication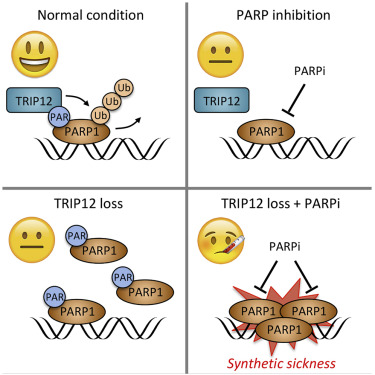
Abstract
PARP inhibitors (PARPi) cause synthetic lethality in BRCA-deficient tumors. Whether specific vulnerabilities to PARPi exist beyond BRCA mutations and related defects in homology-directed repair (HDR) is not well understood. Here, we identify the ubiquitin E3 ligase TRIP12 as negative regulator of PARPi sensitivity. We show that TRIP12 controls steady-state PARP1 levels and limits PARPi-induced cytotoxic PARP1 trapping. Upon loss of TRIP12, elevated PARPi-induced PARP1 trapping causes increased DNA replication stress, DNA damage, cell cycle arrest, and cell death. Mechanistically, we demonstrate that TRIP12 binds PARP1 via a central PAR-binding WWE domain and, using its carboxy-terminal HECT domain, catalyzes polyubiquitylation of PARP1, triggering proteasomal degradation and preventing supra-physiological PARP1 accumulation. Further, in cohorts of breast and ovarian cancer patients, PARP1 abundance is negatively correlated with TRIP12 expression. We thus propose TRIP12 as regulator of PARP1 stability and PARPi-induced PARP trapping, with potential implications for PARPi sensitivity and resistance.
Further reading:
Matteo’s and Federico’s collaborative paper is out
July 2020
Spearheaded by Matteo from Massimo Lopes’ group and Federico from our lab, our collaborative study on RAD51 and replication fork reversal is finally out: Sequential role of RAD51 paralog complexes in replication fork remodeling and restart. Published in Nature Communications.
Read more about the paper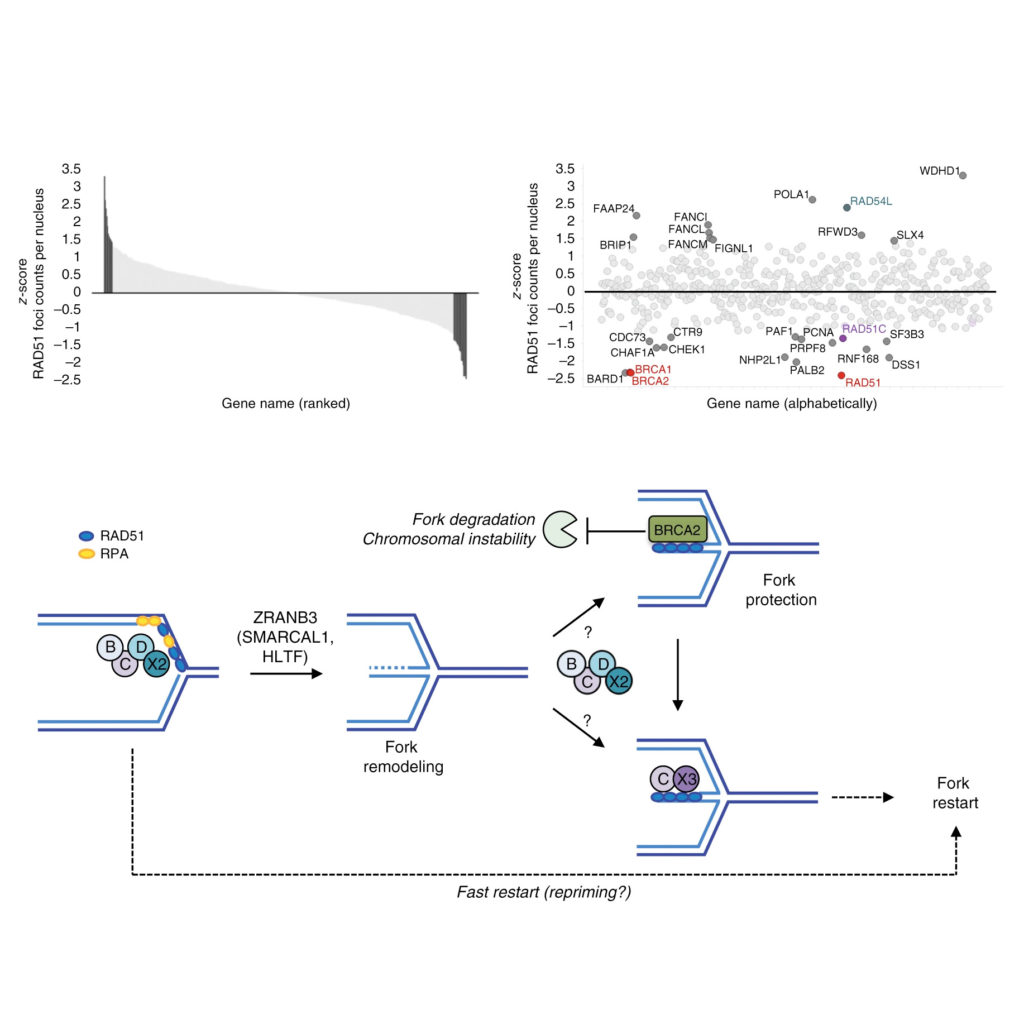
Abstract
Homologous recombination (HR) factors were recently implicated in DNA replication fork remodeling and protection. While maintaining genome stability, HR-mediated fork remodeling promotes cancer chemoresistance, by as-yet elusive mechanisms. Five HR cofactors – the RAD51 paralogs RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3 – recently emerged as crucial tumor suppressors. Albeit extensively characterized in DNA repair, their role in replication has not been addressed systematically. Here, we identify all RAD51 paralogs while screening for modulators of RAD51 recombinase upon replication stress. Single-molecule analysis of fork progression and architecture in isogenic cellular systems shows that the BCDX2 subcomplex restrains fork progression upon stress, promoting fork reversal. Accordingly, BCDX2 primes unscheduled degradation of reversed forks in BRCA2-defective cells, boosting genomic instability. Conversely, the CX3 subcomplex is dispensable for fork reversal, but mediates efficient restart of reversed forks. We propose that RAD51 paralogs sequentially orchestrate clinically relevant transactions at replication forks, cooperatively promoting fork remodeling and restart.
Further reading:
Aswini Krishnan joins the lab as UZH resumes adapted on-site operations
June 2020
Originally planned for beginning of April, Aswini finally obtains permission to enter Switzerland to begin her postdoc fellowship in our group. Not an easy time to start a new research project. With the COVID-19 cases currently being low, restrictions are gradually lifted and UZH resumes on-site operations with applicable safety concepts in place.
Get to know AswiniAswini Krishnan – Postdoctoral Research Scientist

Born and raised in
Kerala, India
Scientific education and research experience
2020 – present: Postdoctoral Research Scientist, University of Zurich, Switzerland
2014 – 2019: PhD Student, Cell Biology Unit, University Medical Center Johannes Gutenberg- University Mainz, Germany
2012 – 2014: Junior Research Fellow, Institute for Stem Cell Biology and Regenerative Medicine, Bangalore, India
2010 – 2012: Scientific Officer, The Beatson Institute for Cancer Research, Glasgow, United Kingdom
2008 – 2009: Master of Science, Stem Cell and Regenerative Medicine, University of Sheffield, United Kingdom
2005 – 2008: Bachelor of Science, Biotechnology, Chemistry, Botany, Bangalore University, India
Research interests
Cancer biology
Translational research
Conferences cancelled, new hires delayed due to COVID-19
May 2020
As the pandemic continues to keep the world in its grip, scientific conferences are cancelled or changed to virtual format. New hires are complicated by restricted international mobility and immigration procedures.
University remains closed in April
April 2020
The University of Zurich remains closed for most on-site activities and operates with skeleton staff. All cells are frozen down, all reagents are stored securely, time to update lab books, catch up with reading & writing, and to stay physically and mentally healthy.
Have a look at the COVID-19 developments in Switzerland from March to May 2020
Source: https://www.corona-data.ch/
University lockdown in response to SARS-CoV-2
March 2020
With the rapid increase in COVID-19 cases in Switzerland and after the first cases at the University of Zurich, all on-site research and teaching activities are suspended. University buildings are closed, online teaching, virtual meetings, and working from home becomes the new normal.
Online lab meetings due to the pandemic
Yanlin Wen joins the lab
February 2020
After finishing her Master thesis at the Max Delbrück Center in Berlin, Germany, and amidst an arising pandemic, Yanlin manages to move from her home country China to Switzerland to start her PhD in our group. Welcome on board!
Get to know YanlinYanlin Wen – PhD Student (Molecular Life Sciences Program)

Born and raised in
China
Scientific Education
2020 – present: Ph.D. student (Molecular Life Science) , University of Zurich, Switzerland
2017 – 2019: M.Sc. Biochemistry, Freie Universität Berlin, Germany
2014 – 2017: B.Sc. Biochemistry, Freie Universität Berlin, Germany
Research interests
DNA replication
Signaling pathways involved in DNA damage response
Replication-recombination-repair (RRR) meeting
February 2020
Emerging concepts and recent findings on replication stress-associated heritable DNA lesions, on specificity and functionality of liquid-liquid phase separation at sites of DNA damage, and on mechanisms of action of small molecule inhibitors targeting DNA damage response proteins towards improved cancer therapy were discussed in our RRR presentations by Aleksandra, Sinan and Marco.
Andreas joins the lab
The DMMD rounds off the year
December 2019
The DMMD Christmas Dinner got everyone together to look back to and celebrate the achievements of the year. Two paper awards were given to our lab in appreciation of the performed work and the great team effort that is behind. Time to say thank you to the Department, the Faculty and UZH for providing opportunities and the framework, infrastructure and leadership to enable this. Merry Christmas everyone!
Graduate Campus Annual Ceremony
November 2019
At the UZH Graduate Campus Annual Ceremony our former PhD student Federico Teloni received the Mercator Award 2019 for his outstanding scientific achievements during his PhD studies. Many thanks to Graduate Campus and the Mercator Foundation Switzerland for a fantastic event and for their great support towards young scientists.
View impressions from the Award Ceremony
First scientific DMMD Retreat
October 2019
For the first time in its history, the Department of Molecular Mechanisms of Disease (DMMD) organized a three-day scientific retreat. Up in Beatenberg with gorgeous views of Eiger, Mönch, Jungfrau, we discussed ongoing research projects and collaborative studies. A big thanks to everyone who contributed to the talks and poster sessions!
View impressions from the retreat
Exciting group summer event in Interlaken
September 2019
To get some fresh air and clear up the minds the group made a day trip to Interlaken in central Switzerland. Unafraid of new challenges and adventures, we went down the local canyon, a great team-building event, which moved us out of our comfort zones.
Group pictures in the canyon
Jone’s project on CHK1 regulation is published in JCB
August 2019
Through a combination of molecular biology and quantitative cell biology, Jone provides evidence that steady-state activity of the cell cycle kinase CHK1 safeguards its own stability to maintain intrinsic checkpoint functions and ensure genome integrity and cell survival. These findings are now out in JCB.
Phase separation of DNA repair compartments in EMBO Journal
July 2019
Our newest article is now out in EMBO Journal. We show that:
- DNA repair compartments show liquid‐like properties with droplet fusions and fissions.
- Phase separation behavior is conferred by 53BP1, and uncoupled from upstream DNA damage detection and γH2AX‐MDC1 accumulation.
- 53BP1 assembly at break sites is abolished by changes in osmotic pressure, temperature, salt concentration, or disruption of hydrophobic interactions.
- 53BP1 optoDroplet experiments indicate multivalency and reveal sequence elements required for clustering.
- Disruption of 53BP1 phase separation impairs p53 activation and p21 induction upon DNA damage.
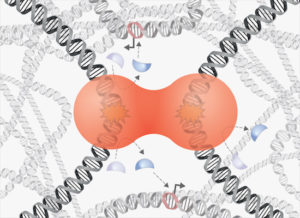
Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments
The DNA damage response (DDR) generates transient repair compartments to concentrate repair proteins and activate signaling factors. The physicochemical properties of these spatially confined compartments and their function remain poorly understood. Here, we establish, based on live cell microscopy and CRISPR/Cas9-mediated endogenous protein tagging, that 53BP1-marked repair compartments are dynamic, show droplet-like behavior, and undergo frequent fusion and fission events. 53BP1 assembly, but not the upstream accumulation of γH2AX and MDC1, is highly sensitive to changes in osmotic pressure, temperature, salt concentration and to disruption of hydrophobic interactions. Phase separation of 53BP1 is substantiated by optoDroplet experiments, which further allowed dissection of the 53BP1 sequence elements that cooperate for light-induced clustering. Moreover, we found the tumor suppressor protein p53 to be enriched within 53BP1 optoDroplets, and conditions that disrupt 53BP1 phase separation impair 53BP1-dependent induction of p53 and diminish p53 target gene expression. We thus suggest that 53BP1 phase separation integrates localized DNA damage recognition and repair factor assembly with global p53-dependent gene activation and cell fate decisions.
Further reading:
Villa Vigoni Meeting in Menaggio
June 2019
With great views on Lake Como, the setting of the Villa Vigoni Meeting on Cell Death and Disease in Menaggio, Italy, made it easy and pleasant to interact and exchange ideas about the past, present and future research on the many flavours of how cells decide to live or die.
PARP 2019 Meeting in Budapest
May 2019
Great presentations, flash talks, and interactive poster sessions at the PARP 2019 Meeting in Budapest, Hungary. A lot of inspiring science on PARP biology, poly(ADP-ribose) functions and PARP inhibitor actions in a fantastic city. Already looking forward to the next edition.
Federico receives prestigious Mercator Award
April 2019
Federico Teloni, who recently defended his Ph.D. work in our lab, receives the prestigious Mercator Award 2019 in the category Natural Sciences & Medicine. The award honours his exciting research findings and outstanding achievements. For further information, please visit:
New protocol on Nature Protocol Exchange
March 2019
We uploaded a detailed protocol on Nature Protocol Exchange to accompany our recent publication in Nature Communictions (Michelena et al. Nat Commun. 2018 Jul 11;9(1):2678). You can read the step-by-step protocol, which also includes a troubleshooting section, here.
Article published in current issue of Molecular Cell
February 2019
Replication stress is a hallmark of many cancers. In an article published in the current issue of Molecular Cell (Volume 73, Issue 4, p641-858) we identify the pre-mRNA cleavage factor WDR33 as regulator of replication stress resilience and demonstrate that, when WDR33 function is impaired, unreleased nascent transcripts and genomic loci re-localize toward the nuclear periphery, where they cause replication stress and DNA damage.
Read more about this work
Efficient Pre-mRNA Cleavage Prevents Replication-Stress-Associated Genome Instability
Cellular mechanisms that safeguard genome integrity are often subverted in cancer. To identify cancer-related genome caretakers, we employed a convergent multi-screening strategy coupled to quantitative image-based cytometry and ranked candidate genes according to multivariate readouts reflecting viability, proliferative capacity, replisome integrity, and DNA damage signaling. This unveiled regulators of replication stress resilience, including components of the pre-mRNA cleavage and polyadenylation complex. We show that deregulation of pre-mRNA cleavage impairs replication fork speed and leads to excessive origin activity, rendering cells highly dependent on ATR function. While excessive formation of RNA:DNA hybrids under these conditions was tightly associated with replication-stress-induced DNA damage, inhibition of transcription rescued fork speed, origin activation, and alleviated replication catastrophe. Uncoupling of pre-mRNA cleavage from co-transcriptional processing and export also protected cells from replication-stress-associated DNA damage, suggesting that pre-mRNA cleavage provides a mechanism to efficiently release nascent transcripts and thereby prevent gene gating-associated genomic instability.
Further reading:
We are on Twitter
January 2019
Follow us on Twitter @altmeyerlab
Edison joins the lab
Stefania successfully defended her PhD thesis
December 2018
Another successful PhD defense! Congratulation to Stefania, who started as the second PhD student of our lab, and who is now the second to have handed in the thesis and passed the final exam. Very well done!!
Impressions from Stefania's PhD celebration
Manuscript accepted & PhD successfully defended
November 2018
What a week, within just a few days our lab member Federico Teloni got his work accepted for publication in Molecular Cell and also successfully defended his PhD thesis. Huge congratulations to him and to the whole lab for these fantastic achievements!
Impressions from Federico's PhD celebration
Vincent joins the lab
Join us at the Science Festival 100 Ways of Thinking
September 2018
Is making music a form of thinking? How many people does it take to think? Why are computers unable to come up with jokes? How does thinking work and what happens when it stops working? Can the universe be represented by a picture, and what is science’s relationship to that picture? Is art more than an imaging process? These are some of the questions posed by 100 Ways of Thinking, taking place from August 25 to November 4, 2018, during which time Kunsthalle Zurich is hosting the University of Zurich.
Read more about the exhibition and the contribution of the DMMD
A Voyage Into The Cell
At the Department of Molecular Mechanisms of Disease (DMMD) cellular and molecular biologists work together to develop various ways of illuminating and understanding the structures and processes that exist inside cells. We work hard to better understand the cell, the smallest subunit of our bodies. If something goes wrong in our cells, the whole body suffers. Numerous diseases are the result of the molecular processes inside our cells somehow being disturbed. Malfunctions that occur at the level of our DNA, our genes, and their regulation can have particularly serious consequences. For example, the cellular metabolism can be disturbed to such an extent so that it develops into diabetes. Mutations in the genetic material can make it easier for cancer to develop. A deregulated response on the part of the immune cells can lead to chronic inflammation. And if gene regulation isn’t working properly, some cells may not even remember what their job is inside the body. Because cells are so small (one cell is about 100 times smaller than a pinhead), biologists need microscopes in order to look at them. Or they use other, sometimes quite complicated, imaging procedures to look inside the cells and watch the molecular mechanisms at work. These procedures are carried out in a research laboratory, they often take several days, and they’re based on a very precise sequence involving numerous steps. If the experiment works, we know a little more about the cell. Based on the new findings, we can then make plans for subsequent experiments. We’ve yet to run out of new questions. Join us at the Science Festival 100 Ways of Thinking at Kunsthalle Zurich from August 25 to November 4, 2018. Entrance is free, so come and explore!
Further information:
Looking back on the DMMD 2018 Summer Games and our group rafting trip
August 2018
Lots of fun games, a proud winning team and a relaxed BBQ made the DMMD 2018 Summer Games a great event. Big thanks to the organizers and everyone for joining! And for the rafting, glad that everyone enjoyed – and survived!
Have a look at some pictures of the 2018 Summer Games and our rafting trip
Measuring cellular responses to PARP inhibition
July 2018
Our newest paper on PARP inhibitor toxicity has been published in Nature Communications! Have a look how high-content imaging reveals the earliest cellular responses to PARP inhibition and unravels the sequence of events from PARP trapping to DNA damage, cell cycle arrest and cell death. Here you can access the complete article and browse through the Editors’ Highlights, or read the UZH/EurekAlert public releases.
Graphical abstract and summary Analysis of PARP inhibitor toxicity by multidimensional fluorescence microscopy reveals mechanisms of sensitivity and resistance
Analysis of PARP inhibitor toxicity by multidimensional fluorescence microscopy reveals mechanisms of sensitivity and resistance
Exploiting the full potential of anti-cancer drugs necessitates a detailed understanding of their cytotoxic effects. While standard omics approaches are limited to cell population averages, emerging single cell techniques currently lack throughput and are not applicable for compound screens. Here, we employed a versatile and sensitive high-content microscopy-based approach to overcome these limitations and quantify multiple parameters of cytotoxicity at the single cell level and in a cell cycle resolved manner. Applied to PARP inhibitors (PARPi) this approach revealed an S-phase-specific DNA damage response after only 15 min, quantitatively differentiated responses to several clinically important PARPi, allowed for cell cycle resolved analyses of PARP trapping, and predicted conditions of PARPi hypersensitivity and resistance. The approach illuminates cellular mechanisms of drug synergism and, through a targeted multivariate screen, could identify a functional interaction between PARPi olaparib and NEDD8/SCF inhibition, which we show is dependent on PARP1 and linked to PARP1 trapping.
Research distinctions for Stefania and Jone
June 2018
Our lab members Stefania and Jone both received distinctions for their work: Stefania won a poster price at the fantastic EMBO Workshop on Chromatin dynamics & nuclear organization in genome maintenance, and Jone received new project funding from the UZH. Very well done!
Approach of the lab featured in BIOspektrum
May 2018
Our research and experimental approach was recently featured in a High Content Imaging (HCI) Special in BIOspektrum, the magazine of the German bioscience societies GBM, VAAM, GfG and DGPT. Have a look at the current issue, HCI is not only about large-scale screens. Here is the article and here the link to the complete issue. Download an English version here.
Editorial on genotoxic stress inheritance
April 2018
Read our short editorial “The memory remains” in Aging on the effects of genotoxic stress inheritance from one cell generation to the next. Access the article via PubMed or the journal homepage.
Lab research featured in UZH Magazin
Up to the mountains to reach for the sky
February 2018
Our traditional winter sledging day took place again high above Elm, eastern Switzerland. Racy descents on a sunny day and a great opportunity to strengthen interactions between the different groups of our department.
View impressions from the slopes
Joint Department Colloquium starts off the year
January 2018
We started into 2018 with our third DMMD Colloquium, a day full of scientific talks and poster sessions to foster discussions and provide an overview of our research activities to newcomers. Special feature this time: an entertaining lecture by communication trainer Dani Nieth.
View impressions from the colloquium
News & Views: Daughters sense their mother’s stress
November 2017
Read the News & Views article on our recently published work on the effects of DNA damage propagation from one cell generation to the next. It nicely puts our work into context with recent findings from the labs of Tobias Meyer, Sabrina Spencer, and Chris Bakal. Access the News & Views via PubMed or the Cell Cycle homepage.
Aleksandra’s paper is out
October 2017
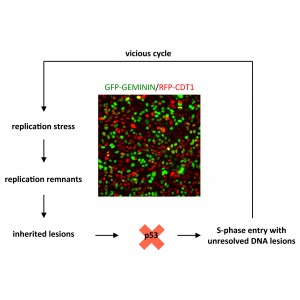
Aleksandra’s work shows how inherited DNA lesions, which originate from replication problems during the previous S-phase, determine G1 duration in daughter cells. Her findings provide a deterministic explanation for cellular heterogeneity in cell cycle commitment and further suggest that mild replication stress during cancer development, induced for instance by oncogene activation, cooperates with loss of tumor suppressor protein p53 to force cells into a “vicious cycle” of damage propagation and accumulation of mutations.
Read moreInherited DNA lesions determine G1 duration in the next cell cycle.
Abstract
Replication stress is a major source of DNA damage and an important driver of cancer development. Replication intermediates that occur upon mild forms of replication stress frequently escape cell cycle checkpoints and can be transmitted through mitosis into the next cell cycle. The consequences of such inherited DNA lesions for cell fate and survival are poorly understood. By using time-lapse microscopy and quantitative image-based cytometry to simultaneously monitor inherited DNA lesions marked by the genome caretaker protein 53BP1 and cell cycle progression, we show that inheritance of 53BP1-marked lesions from the previous S-phase is associated with a prolonged G1 duration in the next cell cycle. These results suggest that cell-to-cell variation in S-phase commitment is determined, at least partially, by the amount of replication-born inherited DNA damage in individual cells. We further show that loss of the tumor suppressor protein p53 overrides replication stress-induced G1 prolongation and allows S-phase entry with excessive amounts of inherited DNA lesions. Thus, replication stress and p53 loss may synergize during cancer development by promoting cell cycle re-entry with unrepaired mutagenic DNA lesions originating from the previous cell cycle.
Link to PubMed:
Inherited DNA lesions determine G1 duration in the next cell cycle.
Cell Cycle. 2017 Oct 5:0. doi: 10.1080/15384101.2017.1383578.
Micheal joins the lab
Joint lab retreat at Flumserberg
September 2017
As they say, never change a winning team: 4 research groups from 3 universities of 2 countries came together for their second joint lab retreat 1378 meters above sea level at Flumserberg Tannenbodenalp, Switzerland. Superb talks, stimulating project discussions, great cooking, a short hike up to a peaceful mountain lake, and an exhilarating toboggan ride back down made this a fantastic group event.
View impressions from the retreat
Poster Prize to lab member Federico Teloni
September 2017
Congratulations to Federico, who won a prize for his poster at the 4th German-French DNA Repair Meeting in Cologne, Germany. Federico presented his project on a convergent multidimensional screen to identify cancer-relevant genes with roles in replication stress resilience. Well done!
Andrew joins the lab
August 2017
Coming from the Friedrich Miescher Institute in Basel, where he obtained his PhD, Andrew Seeber joins the lab as postdoctoral fellow. Supported by the SNF/KTI BRIDGE Program, Andrew will study the role of chromatin dynamics for DSB repair. Welcome on board!
Have a look at Andrew's scientific backgroundAndrew Seeber – SNF BRIDGE Fellow

Born and raised in
South Africa / Ireland
Scientific Education
2017 – present: Postdoctoral Research Scientist, University of Zurich, Switzerland
2011 – 2017: Ph.D. student (Genetics), Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland
2010 – 2011: NCCR Ph.D. program rotations, University of Geneva, Switzerland
2006 – 2010: B.Sc. (Biochemistry), National University of Ireland, Galway, Ireland
Research interests
Chromatin organization and dynamics
DNA double-strand break repair and the DNA damage response with a focus on the role of chromatin modification in these processes
Genome editing
Article finally out in Methods in Molecular Biology
July 2017
Our lab contributed a chapter to the Second Edition of “Poly(ADP-Ribose) Polymerase: Methods and Protocols”, edited by Alexei V. Tulin. Check out how automated microscopy can be used for cell cycle resolved measurements of poly(ADP-ribose) formation.
Read the articleCell Cycle Resolved Measurements of Poly(ADP-Ribose) Formation and DNA Damage Signaling by Quantitative Image-Based Cytometry.
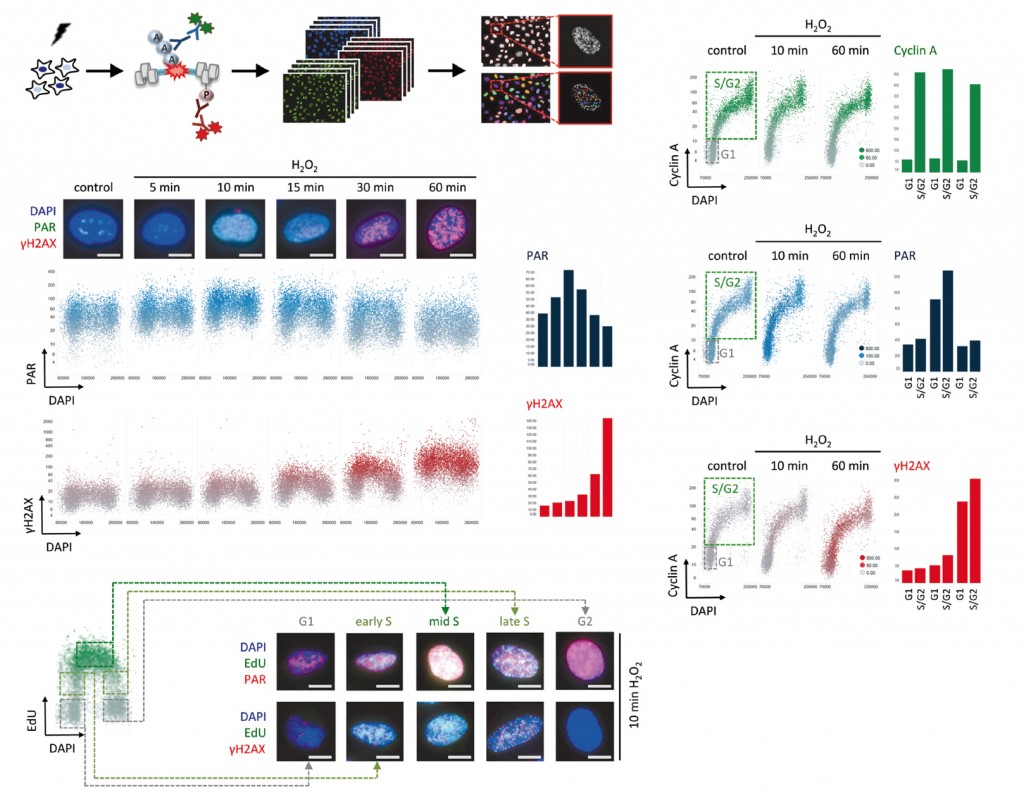
Abstract
Formation of poly(ADP-ribose) (PAR) marks intracellular stress signaling and is notably induced upon DNA damage. PAR polymerases (PARPs) catalyze PAR synthesis upon genotoxic stress and thereby recruit multiple proteins to damaged chromatin. PAR induction is transient and antagonized by the action of PAR glycohydrolase (PARG). Given that poly(ADP-ribosyl)ation (PARylation) is involved in genome integrity maintenance and other vital cellular functions, but also in light of the recent approval of PARP inhibitors for cancer treatments, reliable measurements of intracellular PAR formation have gained importance. Here we provide a detailed protocol for PAR measurements by quantitative image-based cytometry. This technique combines the high spatial resolution of single-cell microscopy with the advantages of cell population measurements through automated high-content imaging. Such upscaling of immunofluorescence-based PAR detection not only increases the robustness of the measurements through averaging across large cell populations but also allows for the discrimination of subpopulations and thus enables multivariate measurements of PAR levels and DNA damage signaling. We illustrate how this technique can be used to assess the dynamics of the cellular response to oxidative damage as well as to PARP inhibitor-induced genotoxicity in a cell cycle resolved manner. Due to the possibility to use any automated microscope for quantitative image-based cytometry, the presented method has widespread applicability in the area of PARP biology and beyond.
Read the full article:
Cell Cycle Resolved Measurements of Poly(ADP-Ribose) Formation and DNA Damage Signaling by Quantitative Image-Based Cytometry.
Methods Mol Biol. 2017;1608:57-68.
Read the complete book:
“Poly(ADP-Ribose) Polymerase: Methods and Protocols”, Second Edition. Edited by Alexei V. Tulin
Methods Mol Biol. Volume 1608 2017. ISBN: 978-1-4939-6992-0 (Print) 978-1-4939-6993-7 (Online)
Worth Repeating: Second Mini-Retreat of the Lab
June 2017
How do cells keep a stable genome? What is the cellular toolkit to repair damaged DNA? How do cells choose which tool to use? And how can quantitative cell biology help tackle these questions? Fresh minds were ready for some brainwork during an inspiring one-day lab retreat.
Snapshots from the retreat
Lab member Jone featured in JoVE
June 2017
Watch our lab member Jone Michelena explain two complementary cell synchronization protocols, which she used during her PhD studies in Ana Zubiaga’s lab in Bilbao, Spain, to study cell cycle regulated gene expression. Her video is now online in JoVE.
Work by Stefania and Jone, Federico and Ralph published in Cell Reports
May 2017
In their work Stefania, Jone, Federico and Ralph show how replication-dependent differences in chromatin inform the DNA damage response machinery about the replication status of broken genomic loci. This chromatin-embedded information affects 53BP1 binding and directs the choice of repair pathway used to fix DNA double-strand breaks.
Read more about our latest work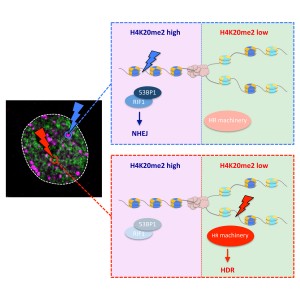
Highlights
Abstract
The bivalent histone modification reader 53BP1 accumulates around DNA double-strand breaks (DSBs), where it dictates repair pathway choice decisions by limiting DNA end resection. How this function is regulated locally and across the cell cycle to channel repair reactions toward non-homologous end joining (NHEJ) in G1 and promote homology-directed repair (HDR) in S/G2 is insufficiently understood. Here, we show that the ability of 53BP1 to accumulate around DSBs declines as cells progress through S phase and reveal that the inverse relationship between 53BP1 recruitment and replicated chromatin is linked to the replication-coupled dilution of 53BP1’s target mark H4K20me2. Consistently, premature maturation of post-replicative chromatin restores H4K20me2 and rescues 53BP1 accumulation on replicated chromatin. The H4K20me2-mediated chromatin association of 53BP1 thus represents an inbuilt mechanism to distinguish DSBs in pre- versus post-replicative chromatin, allowing for localized repair pathway choice decisions based on the availability of replication-generated template strands for HDR.
Read the full article:
Replication-Coupled Dilution of H4K20me2 Guides 53BP1 to Pre-replicative Chromatin
Cell Reports 19, 1819–1831, 2017
Sinan Kilic and Marco Gatti join the lab
Thomas Schmid returns to the lab
April 2017
After finishing his studies in Veterinary Medicine at the Vetsuisse Faculty, Thomas joins our lab again for the experimental part of his dissertation (Dr. med. vet.). Building on his work from the Master’s Thesis, Thomas will study PARP inhibitor toxicity in human cancer cells. Welcome back and good luck!
Joint Department Colloquium and DMMD goes Sledding
February 2017
We started into the year with a Joint Department Colloquium, a day full of inspiring talks, scientific discussions, great poster sessions, snacks & drinks, and games of tabletop soccer. The brainwork was balanced by a day out in the snow sledding down the mountain slopes in Eastern Switzerland.
Have a look at impressions from the 2017 Joint Department Colloquium
Eliana joins the lab
January 2017
Coming from the Department of Biosciences at the University of Milan, Italy, Eliana Bianco was accepted to the Cancer Biology Program of the Life Science Zurich Graduate School and will carry out her PhD project in our lab. Welcome on board and good luck, Eliana!
Have a look at Eliana's scientific backgroundEliana Bianco – PhD Student (Cancer Biology Program)

Born and raised in
Italy
Scientific Education
2017 – present: Ph.D. student (Cancer Biology), University of Zurich, Switzerland
2016: Scholarship for the Promising Young, Laboratory of Genome Instability and Human Pathologies, Department of Bioscience, University of Milan, Italy
2013 – 2015: M.Sc. (Molecular Biotechnology and Bioinformatics), University of Milan, Italy
2010 – 2013: B.Sc. (Biotechnology), University of Milan, Italy
Research interests
Genome instability
DNA repair and DNA damage checkpoints
DNA replication
Postdoc Fellowship awarded to Jone Michelena
December 2016
Our lab member Jone Michelena was awarded a prestigious outgoing postdoctoral fellowship from her home country Spain (through the Gobierno Vasco Programa Posdoctoral de Perfeccionamiento de Personal Investigador Doctor) to support her research in our group. Well done, Jone!
Interview in Vetsuisse News
December 2016
Read a short interview with Matthias Altmeyer and Tuncay Baubec in Vetsuisse News (in German):
Aleksandra joins the lab
November 2016
Coming from Prof. David Shore’s lab at the University of Geneva, Switzerland, Aleksandra Lezaja was accepted to the Cancer Biology Program of the Life Science Zurich Graduate School and will carry out her PhD project in our lab. Welcome on board and good luck, Aleksandra!
Have a look at Aleksandra's scientific backgroundAleksandra Lezaja – PhD Student (Cancer Biology Program)

Born and raised in
Serbia
Scientific Education
2016 – present: Ph.D. student (Cancer Biology), University of Zurich, Switzerland
2014 – 2016: MSc (Biology, Curriculum: Genetics, Development and Evolution), Master Excellence Fellowship, Faculty of Science, University of Geneva, Switzerland
2010 – 2014: BSc (Molecular Biology and Physiology), University of Serbia, Belgrade
Research interests
DNA replication
Signaling pathways involved in DNA damage repair, cancer genetics, cell cycle progression
ERC grant awarded to the lab (ERC-2016-StG)
September 2016
As part of the Horizon 2020 Research Program of the European Union an ERC Starting Grant was awarded to the lab. Open positions for ERC funded projects in our research program will be announced soon. We are extremely grateful for this encouraging support by the European Union.
2016 DMMD Summer Games
July 2016
Our lab joined the 2016 DMMD Summer Games. Lots of fun games, a proud winning team, an improvised soccer match against young physicist Olympians from Egypt, and a relaxed BBQ made this a great event. Big thanks to the organizers and everyone for joining!
Have a look at the 2016 Summer Games Department group photo
Additional research support for the lab
July 2016
We are grateful for financial support by the Novartis Foundation for Medical-Biological Research and by the Swiss Foundation to Combat Cancer (Stiftung zur Krebsbekämpfung). Visit their websites:
PhD Fellowship awarded to Federico Teloni
June 2016
We are proud to announce that our lab member Federico Teloni was awarded the prestigious Candoc PhD Fellowship from the University of Zurich for his project “Defining cancer-specific alterations in genome integrity maintenance”. Well done and all the best for the project!
Joint lab retreat at Flumserberg
June 2016
4 research groups from 3 universities of 2 countries come together for a joint lab retreat 1378 meters above sea level at Flumserberg Tannenbodenalp, Switzerland. Superb talks, stimulating project discussions, great cooking, a BBQ in the rain, and a short hike up to a peaceful mountain lake made this a fantastic event.
View impressions from the retreat
Pakize joins the lab
June 2016
After obtaining her PhD from Ege University in Izmir Pakize became Assistant Professor at Cumhuriyet University in Sivas, Turkey. Supported by the Scientific and Technological Research Council of Turkey she now joined our lab as guest scientist. Welcome and good luck for the project!
Have a look at Pakize's scientific backgroundPakize Canturk – Visiting Research Scientist

Born and raised in
Turkey
Scientific Education
2016 – present: Visiting Research Scientist, University of Zurich, Switzerland
2013 – present: Faculty position, Cumhuriyet University, Sivas, Turkey
2008 – 2012: Ph.D. (Biotechnology), Ege University, Izmir, Turkey
2005 – 2008: M.Sc. (Pharmaceutical Biotechnology), Ege University, Izmir, Turkey
2000 – 2005: B.Sc. (Biology), Ege University, Izmir, Turkey
Research interests
Roles of DNA topoisomerases in anticancer drug mechanisms and replication stress.
Mini-Review published in Frontiers in Genetics
May 2016
We contributed a mini-review to a Frontiers in Genetics research topic dedicated to “Ubiquitin and Ubiquitin-relative SUMO in DNA damage response”. Our article on “Interplay between Ubiquitin, SUMO, and Poly(ADP-Ribose) in the Cellular Response to Genotoxic Stress” is open-access and freely available.
Read more about chain-like protein modifications and how they regulate the cellular responses to genotoxic stressInterplay between Ubiquitin, SUMO, and Poly(ADP-Ribose) in the Cellular Response to Genotoxic Stress
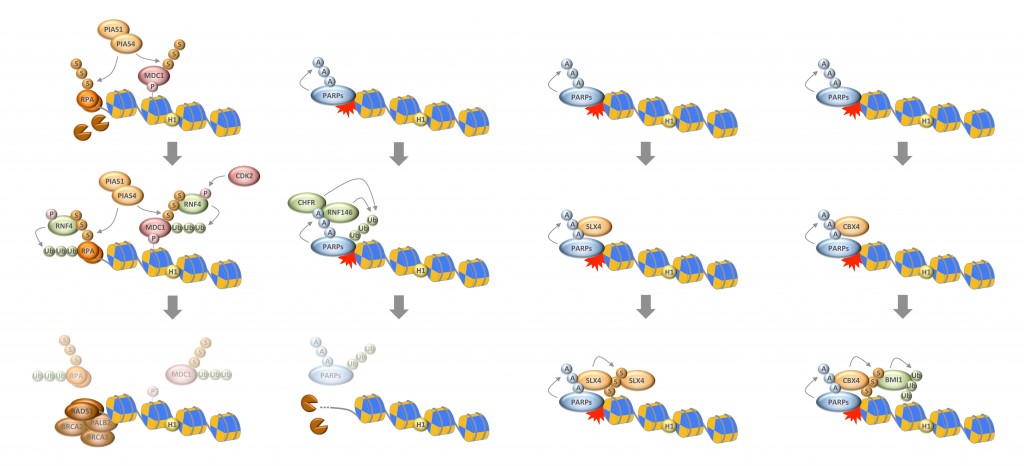
Cells employ a complex network of molecular pathways to cope with endogenous and exogenous genotoxic stress. This multilayered response ensures that genomic lesions are efficiently detected and faithfully repaired in order to safeguard genome integrity. The molecular choreography at sites of DNA damage relies heavily on post-translational modifications (PTMs). Protein modifications with ubiquitin and the small ubiquitin-like modifier SUMO have recently emerged as important regulatory means to coordinate DNA damage signaling and repair. Both ubiquitylation and SUMOylation can lead to extensive chain-like protein modifications, a feature that is shared with yet another DNA damage-induced PTM, the modification of proteins with poly(ADP-ribose) (PAR). Chains of ubiquitin, SUMO, and PAR all contribute to the multi-protein assemblies found at sites of DNA damage and regulate their spatio-temporal dynamics. Here, we review recent advancements in our understanding of how ubiquitin, SUMO and PAR coordinate the DNA damage response and highlight emerging examples of an intricate interplay between these chain-like modifications during the cellular response to genotoxic stress.
Read the full article:
Interplay between Ubiquitin, SUMO, and Poly(ADP-Ribose) in the Cellular Response to Genotoxic Stress
Front. Genet., 19 April 2016 | http://dx.doi.org/10.3389/fgene.2016.00063
Link to the research topic:
Ubiquitin and Ubiquitin-relative SUMO in DNA damage response
Front. Genet., 2016
Review on phase separation out in Trends in Cell Biology
April 2016
Phase separation has emerged as an important means to compartmentalize the intracellular space. We are proud to present our newest review, in which we discuss regulatory principles and physiological functions of phase separation and point to intriguing links to neurodegeneration and cancer.
Read more about (patho)physiological transitions from liquid to solid protein statesPhase separation: linking cellular compartmentalization to disease
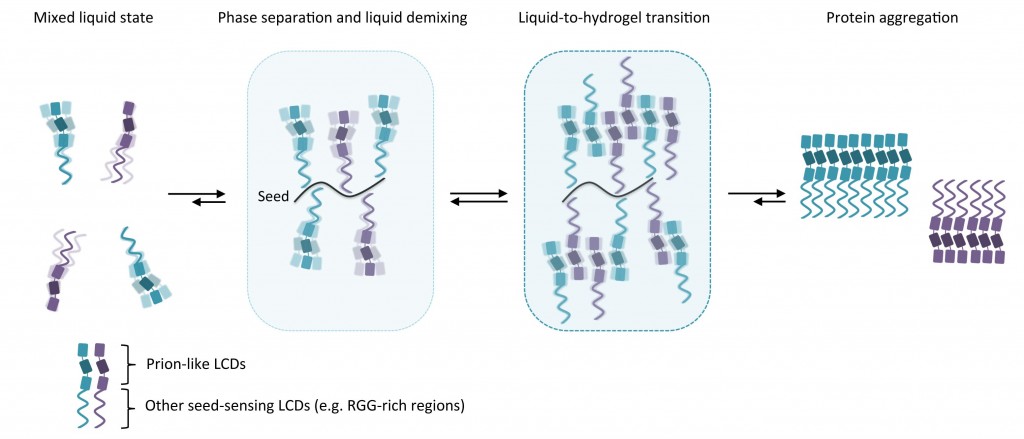
Eukaryotic cells are complex structures capable of coordinating numerous biochemical reactions in space and time. Key to such coordination is the subdivision of intracellular space into functional compartments. Compartmentalization can be achieved by intracellular membranes, which surround organelles and act as physical barriers. In addition, cells have developed sophisticated mechanisms to partition their inner substance in a tightly regulated manner. Recent studies provide compelling evidence that membraneless compartmentalization can be achieved by liquid demixing, a process culminating in liquid–liquid phase separation and the formation of phase boundaries. In this review, we discuss how this emerging concept may help in understanding dynamic reorganization of subcellular space and highlight its potential as a framework to explain pathological protein assembly in cancer and neurodegeneration.
Read the full article:
Phase Separation: Linking Cellular Compartmentalization to Disease
Trends in Cell Biol. 2016 10.1016/j.tcb.2016.03.004
New publications by our lab members Jone and Federico
March/April 2016
Proud to announce that Jone published her PhD work conducted at the University of the Basque Country (Bilbao, Spain) on cell proliferation control by E2F7-regulated microRNAs in Nucleic Acids Research and that Federico’s Master’s Thesis at the University of Michigan (Ann Arbor, USA) made an important contribution to a study just published in Nature Communications.
When time is too short: How ESCs deal with stress
February 2016
A new paper from the Lopes lab with contributions from our group demonstrates that signatures of replication stress in embryonic stem cells (ESCs) can be suppressed by prolonging the G1 phase, indicating that when time in G1 is scarce, ESCs employ replication-coupled mechanisms to maintain genome integrity.
Read more about replication stress in ESCsA short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells.
Embryonic stem cells (ESCs) represent a transient biological state, where pluripotency is coupled with fast proliferation. ESCs display a constitutively active DNA damage response (DDR), but its molecular determinants have remained elusive. Here we show in cultured ESCs and mouse embryos that H2AX phosphorylation is dependent on Ataxia telangiectasia and Rad3 related (ATR) and is associated with chromatin loading of the ssDNA-binding proteins RPA and RAD51. Single-molecule analysis of replication intermediates reveals massive ssDNA gap accumulation, reduced fork speed and frequent fork reversal. All these marks of replication stress do not impair the mitotic process and are rapidly lost at differentiation onset. Delaying the G1/S transition in ESCs allows formation of 53BP1 nuclear bodies and suppresses ssDNA accumulation, fork slowing and reversal in the following S-phase. Genetic inactivation of fork slowing and reversal leads to chromosomal breakage in unperturbed ESCs. We propose that rapid cell cycle progression makes ESCs dependent on effective replication-coupled mechanisms to protect genome integrity.
Read the full article:
A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells.
Nat Commun. 2016 Feb 15;7:10660. doi: 10.1038/ncomms10660.
Sledding day in Elm
January 2016
An opportunity to interact with other members of the institute and discuss science in a relaxed atmosphere, and an opportunity to appreciate the beauty of nature and the magnificence of the Swiss Alps. No broken bones.
View impressions from the day out in the snow
Jone joins the lab
January 2016
Coming from the lab of Prof. Ana Zubiaga at the University of the Basque Country UPV/EHU, Spain, Jone Michelena recently joined our group as a postdoctoral research scientist to strengthen the team. Welcome on board and good luck, Jone!
Have a look at Jone's scientific backgroundJone Michelena – Postdoctoral Research Scientist

Born and raised in
Spain
Scientific Education
2016 – present: Postdoctoral Research Scientist, University of Zurich, Switzerland
2014 – 2015: Postdoctoral Research Scientist, University of the Basque Country UPV/EHU, Spain
2009 – 2014: Ph.D. student (Molecular Biology and Biomedicine), University of the Basque Country UPV/EHU, Spain
2008 – 2009: M.Sc. (Molecular Biology and Biomedicine), University of the Basque Country UPV/EHU and University of Cantabria, Spain
2003 – 2008: B.Sc. (Biochemistry), University of the Basque Country UPV/EHU, Spain
Research interests
Cell cycle, DNA replication and genome stability
Gene expression regulation in response to DNA damage
IVBMB is now Department of Molecular Mechanisms of Disease
January 2016
To better reflect its scientific goals and research activities, our host institute has received a new name and is now the Department of Molecular Mechanisms of Disease (DMMD), associated with both the Science Faculty and the Vetsuisse Faculty of the University of Zurich, Switzerland.
New review article published
December 2015
In this review we provide an update on the growing number of readers of poly(ADP-ribose) and discuss their extraordinary structural and functional diversity. Our article has been published in Nucleic Acids Research.
Read more about the diversity of PAR readersReaders of poly(ADP-ribose): designed to be fit for purpose
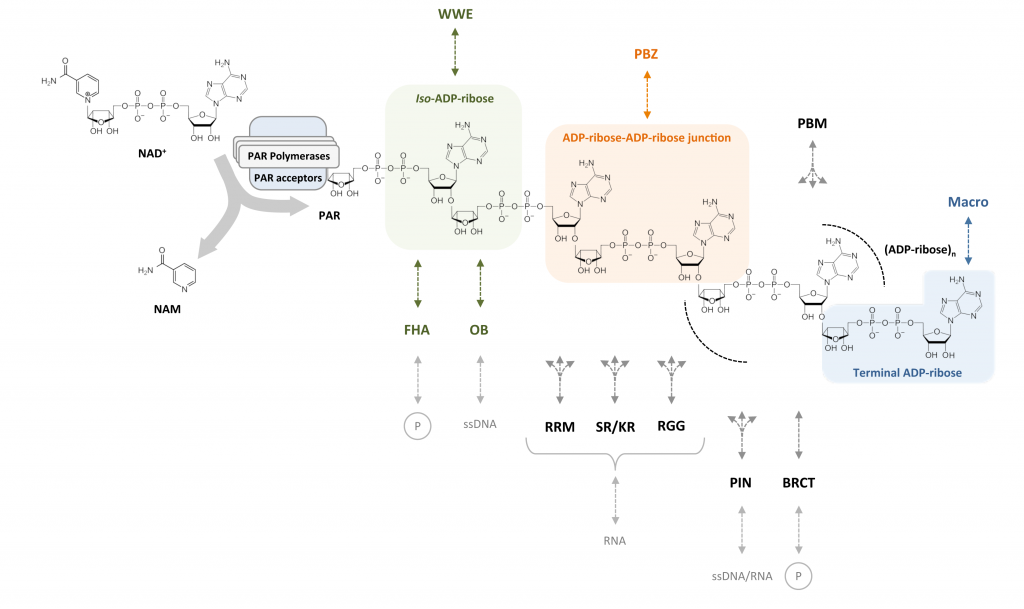
Post-translational modifications (PTMs) regulate many aspects of protein function and are indispensable for the spatio-temporal regulation of cellular processes. The proteome-wide identification of PTM targets has made significant progress in recent years, as has the characterization of their writers, readers, modifiers and erasers. One of the most elusive PTMs is poly(ADP-ribosyl)ation (PARylation), a nucleic acid-like PTM involved in chromatin dynamics, genome stability maintenance, transcription, cell metabolism and development. In this article, we provide an overview on our current understanding of the writers of this modification and their targets, as well as the enzymes that degrade and thereby modify and erase poly(ADP-ribose) (PAR). Since many cellular functions of PARylation are exerted through dynamic interactions of PAR-binding proteins with PAR, we discuss the readers of this modification and provide a synthesis of recent findings, which suggest that multiple structurally highly diverse reader modules, ranging from completely folded PAR-binding domains to intrinsically disordered sequence stretches, evolved as PAR effectors to carry out specific cellular functions.
Read the full article:
Readers of poly(ADP-ribose): designed to be fit for purpose
Nucleic Acids Res. 2016 Feb 18;44(3):993-1006.
First scientific lab retreat
October 2015
Plenty of time to discuss science, ongoing research projects in the lab and our future plans. Worth to repeat.
View impressions from the retreat
New research article accepted for publication
July 2015
In a collaboration with the lab of Jiri Lukas in Copenhagen we discovered that poly(ADP-ribose) functions as nucleation event to initiate liquid-liquid phase separation of intrinsically disordered proteins at sites of DNA damage. Our article has been published in Nature Communications.
Read more about PAR-seeded phase separationsLiquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose)
 Compartmentalization is key to sub-divide the intracellular environment and orchestrate biochemical reactions in space and time. How compartmentalization is achieved is incompletely understood, in particular when it occurs without membranes as physical barriers.
Compartmentalization is key to sub-divide the intracellular environment and orchestrate biochemical reactions in space and time. How compartmentalization is achieved is incompletely understood, in particular when it occurs without membranes as physical barriers.
In collaboration with the group of Prof. Jiri Lukas in Copenhagen, Denmark, we recently found that, in response to DNA damage, membrane-less compartmentalization is achieved by the regulated phase separation of intrinsically disordered proteins. This liquid demixing is initiated by poly(ADP-ribose) (PAR), a nucleic acid-like polymer that is induced at DNA break sites and acts as nucleation event for the rapid and reversible assembly of various unstructured, aggregation-prone proteins. The dynamic compartmentalization through PAR-seeded liquid demixing orchestrates the recruitment kinetics of repair proteins and shields the broken DNA from unwanted reactions.
The study, which was published on August 19th 2015 in Nature Communications, suggests that PAR-seeded liquid demixing is a general mechanism to dynamically reorganize the soluble nuclear space. In light of the aggregation-prone nature of the PAR-responsive intrinsically disordered proteins, deregulation of PAR-seeded liquid demixing may have important implications for pathological protein aggregation during neurodegeneration and aging.
Further reading:
Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose)
Nature Comm. 2015 Aug 19;6:8088
PAR and the organization of the DNA damage response
Nat Struct Mol Biol. 2015 Sep;22(9):655.
UZH News
Our group joins the Hottiger and Baubec labs for a kart race
July 2015
This year’s social summer event was dedicated to kart racing together with the labs of Michael Hottiger and Tuncay Baubec followed by a joint dinner.
View impressions from the race
Matthias receives research grant from the Research Talent Development Fund
April 2015
Significant research funding will be provided by the Research Talent Development Fund (Fonds zur Förderung des akademischen Nachwuchses, FAN). We are very grateful for the trust in our work and the financial support of our research by the FAN.
Thomas joins the lab
March 2015
Thomas Schmid, student of Veterinary Medicine at the University of Zurich, will carry out his Master´s Thesis in our lab. Welcome on board and good luck, Thomas!
Get to know ThomasThomas Schmid – Master Student (Vet Med)

Stefania joins the lab
November 2014
Coming from the Laboratory of Genome Instability and Nuclear Organization headed by Karine Dubrana at the CEA in Fontenay-Aux-Roses, France, Stefania Pellegrino was accepted to the Molecular Life Sciences Program of the Life Science Zurich Graduate School and will carry out her PhD project in our lab. Welcome on board and good luck, Stefania!
Have a look at Stefania's scientific backgroundStefania Pellegrino – PhD Student (MLS)

Born and raised in
Italy
Scientific Education
2014 – present: Ph.D. student (Molecular Biology), University of Zurich, Switzerland
2014: Leonardo Da Vinci scholarship, Laboratory of Genetic Instability and Nuclear Organization, CEA, Fontenay-aux-roses, France
2013 – 2014: Master of II level in “Molecular diagnostics and Translational Biomedicine”, University of Catania, Italy
2010 – 2012: MSc (Cellular and Molecular Biology), University of Catania, Italy
2005 – 2010: BSc (Biology), University of Catania, Italy
Research interests
Genome instability and DNA repair in cancer
Microscopy
Federico joins the lab
October 2014
Following research training abroad in the Department of Cell and Developmental Biology at the University of Michigan in Ann Arbor, USA, Federico Teloni was accepted to the Molecular Life Sciences Program of the Life Science Zurich Graduate School and will carry out his PhD project in our lab. Welcome on board and good luck, Federico!
Have a look at Federico's scientific backgroundFederico Teloni – PhD Student (MLS)

Born and raised in
Italy
Scientific Education
2014 – present: Ph.D. student (Molecular Biology), University of Zurich, Switzerland
2013 – 2014: Scholarship for Abroad Training, University of Michigan, United States of America
2010 – 2013: MSc (Biology, Curriculum: Molecular Diagnostics and Biotechnology), University of Camerino, Italy
2006 – 2010: BSc (Biotechnology), University of Camerino, Italy
Research interests
Genome instability and DNA repair in cancer
Microscopy techniques
Cell biology
Ralph joins the lab
October 2014
Ralph Imhof, an experienced research technician and lab manager and a long-standing member of the institute, joins the lab. His scientific and organizational support will be invaluable for a successful start of our research group. Welcome on board, Ralph!
Have a look at Ralph's scientific backgroundRalph Imhof – Research Technician

Born and raised in
Switzerland
Scientific Education
2007 – present: Lab Technician mbA, IT-Management & Support, Financial Controlling, University of Zurich, Switzerland
2005 – 2007: Lab Manager & new Lab Establishment, Donnelly Centre for Cellular & Biomolecular Research, University of Toronto, Dept. of Biochemistry & Dept. of Molecular Genetics, Canada
1998 – 2005: Lab Technician, IT-Management & Support, Institute for Veterinary Biochemistry and Molecular Biology, University of Zurich, Switzerland
Research interests
Protein Purification & Protein Interaction Methodologies
Molecular Biology
Analytical & preparative HPLC
Matthias joins the University of Zurich
October 2014
With the start of his SNF Professorship in October 2014, Matthias joins the University of Zurich to establish his research group at the IVBMB.
Have a look at Matthias' scientific backgroundMatthias Altmeyer – Research Group Leader

Born and raised in
Germany
Scientific Education
2014 – present: SNF Professor, Principal Investigator, University of Zurich, Switzerland
2012 – 2014: Postdoctoral Research Scientist, Novo Nordisk Foundation Center for Protein Research, Copenhagen, Denmark
2010 – 2012: Postdoctoral Research Scientist, Danish Cancer Society, Copenhagen, Denmark
2007 – 2010: Ph.D. Candidate (Molecular Biology), University of Zurich, Switzerland
2001 – 2006: Diploma Candidate (Biology), University of Konstanz, Germany
Research interests
Genome instability in cancer and aging
Chromatin modifications in response to DNA damage
Cancer-specific alterations in the cellular DNA repair network and their therapeutic potential
Matthias receives SNF Professorship
February 2014
Following his postdoctoral research training first at the Danish Cancer Society and then at the Novo Nordisk Foundation Center for Protein Research in Copenhagen, Denmark, Matthias Altmeyer was awarded an SNF Professorship to be carried out at the University of Zurich. This generous start-up package provided by the Swiss National Science Foundation will allow for the establishment of a new research group dedicated to elucidating molecular mechanisms of genome integrity maintenance. Read more about research funding by the SNF.
